Decoding Protein Folding and Digestion via Diffusion MP-SPR
Application of MP-SPR for Protein Conformation and Fragmentation Kinetics
A novel application of Multi-Parametric Surface Plasmon Resonance (MP-SPR) has been developed to investigate protein folding states and enzymatic degradation through a diffusion-based approach. This technique enables label-free, real-time determination of diffusion coefficients (D) for proteins and small molecules in solution, providing new insights into conformational changes and proteolytic fragmentation.
In this study, bovine serum albumin (BSA) was used as a model protein to investigate structural transitions under different pH and surfactant (SDS) conditions. MP-SPR measurements were performed using a microfluidic setup where BSA solutions were injected into a laminar flow channel above a gold sensor surface. Variations in the SPR angle as a function of time were recorded as the molecules diffused across a sharp concentration interface.
To derive diffusion coefficients from the MP-SPR data, the first derivative of the SPR angle versus time (Δθ/Δt) was computed. The resulting curves were then matched to a large library of simulated diffusion profiles using Gillespie’s stochastic algorithm and quantitatively compared using the Discrete Fréchet distance (dF). This curve-matching approach enables the extraction of the D value that best represents the experimental diffusion behavior. For further details, see the original publication.
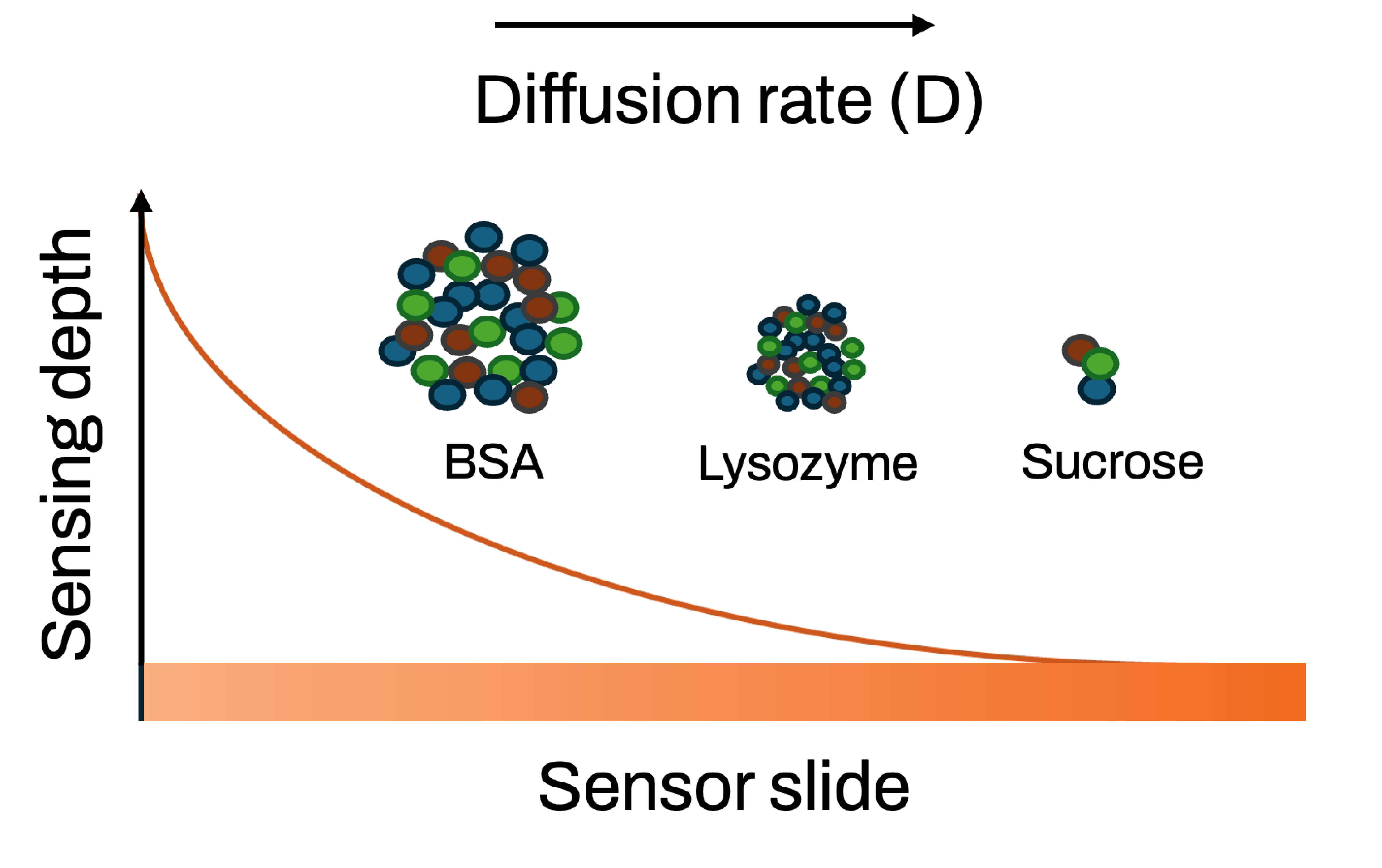
The method was validated using a range of small molecules (e.g., ethanol, sucrose) and extended to larger macromolecules like lysozyme and BSA. Importantly, BSA diffusion was shown to change with its folding state: native, expanded (acidic pH), and denatured (SDS-treated) forms all exhibited distinct D values. Fluorescence spectroscopy confirmed these conformational states.
Furthermore, Diffusion MP-SPR was applied to monitor real-time proteolytic digestion of BSA by trypsin. Over 22 hours, the MP-SPR curves revealed a progressive shift to higher D values, corresponding to the breakdown of the protein into smaller peptide fragments. This label-free, semi-quantitative and kinetic tracking highlights Diffusion MP-SPR’s potential in fragmentomics and enzymology.
By incorporating sampling depth effects, Taylor dispersion theory, and parabolic flow profiles, the study demonstrates that MP-SPR can accurately characterize diffusion phenomena for small molecules and proteins.
To enable accurate diffusion monitoring, the MP-SPR instrument from BioNavis was equipped with custom long injection tubing. If you find this application interesting, please contact info@bionavis.com for further information and guidance on how to apply this to your instrument/ research.
Original publication: Calcagno et al., Chemistry‐Methods, 2025: e202400034.